
Structural model: one-compartment, single bolus IV dose, parameterized using CL driving an EMAX model with a direct effect.
Source:R/models.R
ff.PKPD.1.comp.sd.CL.emax.RdThis is a structural model function that encodes the model described above.
The function is suitable for input to the create.poped.database function using the
ff_fun or ff_file argument.
Arguments
- model_switch
a vector of values, the same size as
xt, identifying which model response should be computed for the corresponding xt value. Used for multiple response models.- xt
a vector of independent variable values (often time).
- parameters
A named list of parameter values.
- poped.db
a poped database. This can be used to extract information that may be needed in the model file.
Value
A list consisting of:
y the values of the model at the specified points.
poped.db A (potentially modified) poped database.
See also
Other models:
feps.add(),
feps.add.prop(),
feps.prop(),
ff.PK.1.comp.oral.md.CL(),
ff.PK.1.comp.oral.md.KE(),
ff.PK.1.comp.oral.sd.CL(),
ff.PK.1.comp.oral.sd.KE(),
ff.PKPD.1.comp.oral.md.CL.imax()
Other structural_models:
ff.PK.1.comp.oral.md.CL(),
ff.PK.1.comp.oral.md.KE(),
ff.PK.1.comp.oral.sd.CL(),
ff.PK.1.comp.oral.sd.KE(),
ff.PKPD.1.comp.oral.md.CL.imax()
Examples
library(PopED)
## find the parameters that are needed to define from the structural model
ff.PKPD.1.comp.sd.CL.emax
#> function (model_switch, xt, parameters, poped.db)
#> {
#> with(as.list(parameters), {
#> y = xt
#> MS <- model_switch
#> CONC = DOSE/V * exp(-CL/V * xt)
#> EFF = E0 + CONC * EMAX/(EC50 + CONC)
#> y[MS == 1] = CONC[MS == 1]
#> y[MS == 2] = EFF[MS == 2]
#> return(list(y = y, poped.db = poped.db))
#> })
#> }
#> <bytecode: 0x55707c2d3df0>
#> <environment: namespace:PopED>
## -- parameter definition function
## -- names match parameters in function ff
sfg <- function(x,a,bpop,b,bocc){
## -- parameter definition function
parameters=c(
CL=bpop[1]*exp(b[1]) ,
V=bpop[2]*exp(b[2]) ,
E0=bpop[3]*exp(b[3]) ,
EMAX=bpop[4]*exp(b[4]) ,
EC50=bpop[5]*exp(b[5]) ,
DOSE=a[1]
)
return( parameters )
}
feps <- function(model_switch,xt,parameters,epsi,poped.db){
## -- Residual Error function
## -- Proportional PK + additive PD
returnArgs <- do.call(poped.db$model$ff_pointer,list(model_switch,xt,parameters,poped.db))
y <- returnArgs[[1]]
poped.db <- returnArgs[[2]]
MS <- model_switch
prop.err <- y*(1+epsi[,1])
add.err <- y+epsi[,2]
y[MS==1] = prop.err[MS==1]
y[MS==2] = add.err[MS==2]
return(list( y= y,poped.db =poped.db ))
}
## -- Define initial design and design space
poped.db <- create.poped.database(ff_fun=ff.PKPD.1.comp.sd.CL.emax,
fError_fun=feps,
fg_fun=sfg,
groupsize=20,
m=3,
sigma=diag(c(0.15,0.15)),
bpop=c(CL=0.5,V=0.2,E0=1,EMAX=1,EC50=1),
d=c(CL=0.01,V=0.01,E0=0.01,EMAX=0.01,EC50=0.01),
xt=c( 0.33,0.66,0.9,5,0.1,1,2,5),
model_switch=c( 1,1,1,1,2,2,2,2),
minxt=0,
maxxt=5,
a=rbind(2.75,5,10),
bUseGrouped_xt=1,
maxa=10,
mina=0.1)
## create plot of model without variability
plot_model_prediction(poped.db,facet_scales="free")
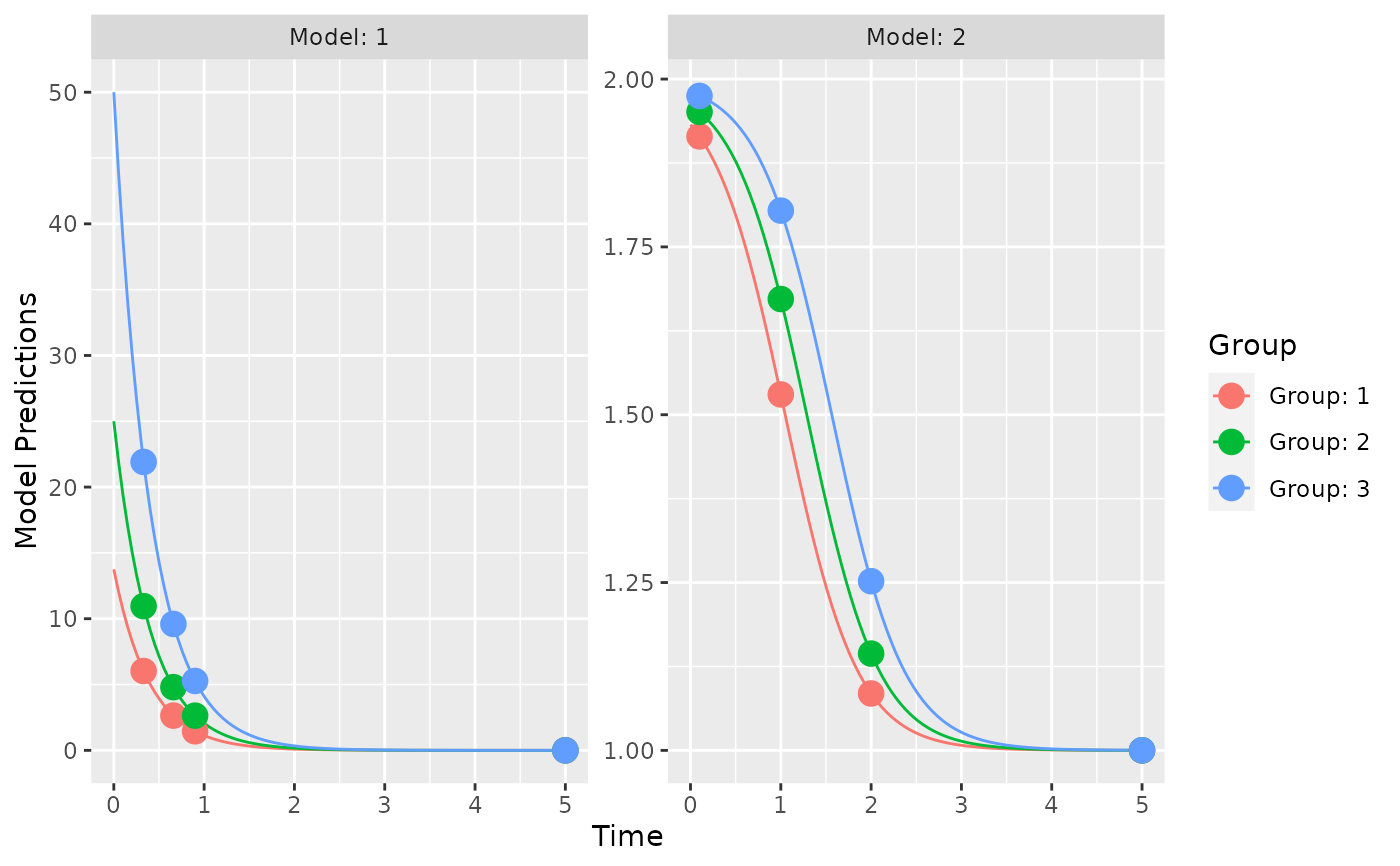 ## evaluate initial design
FIM <- evaluate.fim(poped.db)
FIM
#> [,1] [,2] [,3] [,4] [,5] [,6]
#> [1,] 13423.83149 -26148.43438 -104.9746 -55.87794 20.58619 0.000000e+00
#> [2,] -26148.43438 70314.08833 -137.2793 -103.59890 26.32348 0.000000e+00
#> [3,] -104.97460 -137.27932 1209.7986 516.06655 -116.00915 0.000000e+00
#> [4,] -55.87794 -103.59890 516.0666 449.35861 -62.47406 0.000000e+00
#> [5,] 20.58619 26.32348 -116.0092 -62.47406 22.73598 0.000000e+00
#> [6,] 0.00000 0.00000 0.0000 0.00000 0.00000 9.385378e+04
#> [7,] 0.00000 0.00000 0.0000 0.00000 0.00000 5.697839e+04
#> [8,] 0.00000 0.00000 0.0000 0.00000 0.00000 2.298710e+01
#> [9,] 0.00000 0.00000 0.0000 0.00000 0.00000 6.524561e+00
#> [10,] 0.00000 0.00000 0.0000 0.00000 0.00000 8.880407e-01
#> [11,] 0.00000 0.00000 0.0000 0.00000 0.00000 1.115568e+03
#> [12,] 0.00000 0.00000 0.0000 0.00000 0.00000 1.345372e+01
#> [,7] [,8] [,9] [,10] [,11]
#> [1,] 0.000000e+00 0.000000 0.0000000 0.0000000 0.0000000
#> [2,] 0.000000e+00 0.000000 0.0000000 0.0000000 0.0000000
#> [3,] 0.000000e+00 0.000000 0.0000000 0.0000000 0.0000000
#> [4,] 0.000000e+00 0.000000 0.0000000 0.0000000 0.0000000
#> [5,] 0.000000e+00 0.000000 0.0000000 0.0000000 0.0000000
#> [6,] 5.697839e+04 22.987097 6.5245606 0.8880407 1115.5680152
#> [7,] 6.592095e+04 6.586597 3.6779636 0.2558559 1176.6041001
#> [8,] 6.586597e+00 12197.738218 2243.7358555 112.3394149 4.8902658
#> [9,] 3.677964e+00 2243.735855 1714.4795989 32.6395909 0.5891989
#> [10,] 2.558559e-01 112.339415 32.6395909 4.3373414 0.1964359
#> [11,] 1.176604e+03 4.890266 0.5891989 0.1964359 3811.1481605
#> [12,] 4.616968e+00 3055.546022 1230.5356715 65.5594938 3.8335502
#> [,12]
#> [1,] 0.000000
#> [2,] 0.000000
#> [3,] 0.000000
#> [4,] 0.000000
#> [5,] 0.000000
#> [6,] 13.453723
#> [7,] 4.616968
#> [8,] 3055.546022
#> [9,] 1230.535671
#> [10,] 65.559494
#> [11,] 3.833550
#> [12,] 4658.810828
det(FIM)
#> [1] 1.421054e+39
get_rse(FIM,poped.db)
#> CL V E0 EMAX EC50 d_CL
#> 3.306524 3.610227 4.575451 6.825531 30.397923 47.350220
#> d_V d_E0 d_EMAX d_EC50 SIGMA[1,1] SIGMA[2,2]
#> 56.556024 113.036005 291.542904 5854.936456 10.829134 11.705173
## evaluate initial design
FIM <- evaluate.fim(poped.db)
FIM
#> [,1] [,2] [,3] [,4] [,5] [,6]
#> [1,] 13423.83149 -26148.43438 -104.9746 -55.87794 20.58619 0.000000e+00
#> [2,] -26148.43438 70314.08833 -137.2793 -103.59890 26.32348 0.000000e+00
#> [3,] -104.97460 -137.27932 1209.7986 516.06655 -116.00915 0.000000e+00
#> [4,] -55.87794 -103.59890 516.0666 449.35861 -62.47406 0.000000e+00
#> [5,] 20.58619 26.32348 -116.0092 -62.47406 22.73598 0.000000e+00
#> [6,] 0.00000 0.00000 0.0000 0.00000 0.00000 9.385378e+04
#> [7,] 0.00000 0.00000 0.0000 0.00000 0.00000 5.697839e+04
#> [8,] 0.00000 0.00000 0.0000 0.00000 0.00000 2.298710e+01
#> [9,] 0.00000 0.00000 0.0000 0.00000 0.00000 6.524561e+00
#> [10,] 0.00000 0.00000 0.0000 0.00000 0.00000 8.880407e-01
#> [11,] 0.00000 0.00000 0.0000 0.00000 0.00000 1.115568e+03
#> [12,] 0.00000 0.00000 0.0000 0.00000 0.00000 1.345372e+01
#> [,7] [,8] [,9] [,10] [,11]
#> [1,] 0.000000e+00 0.000000 0.0000000 0.0000000 0.0000000
#> [2,] 0.000000e+00 0.000000 0.0000000 0.0000000 0.0000000
#> [3,] 0.000000e+00 0.000000 0.0000000 0.0000000 0.0000000
#> [4,] 0.000000e+00 0.000000 0.0000000 0.0000000 0.0000000
#> [5,] 0.000000e+00 0.000000 0.0000000 0.0000000 0.0000000
#> [6,] 5.697839e+04 22.987097 6.5245606 0.8880407 1115.5680152
#> [7,] 6.592095e+04 6.586597 3.6779636 0.2558559 1176.6041001
#> [8,] 6.586597e+00 12197.738218 2243.7358555 112.3394149 4.8902658
#> [9,] 3.677964e+00 2243.735855 1714.4795989 32.6395909 0.5891989
#> [10,] 2.558559e-01 112.339415 32.6395909 4.3373414 0.1964359
#> [11,] 1.176604e+03 4.890266 0.5891989 0.1964359 3811.1481605
#> [12,] 4.616968e+00 3055.546022 1230.5356715 65.5594938 3.8335502
#> [,12]
#> [1,] 0.000000
#> [2,] 0.000000
#> [3,] 0.000000
#> [4,] 0.000000
#> [5,] 0.000000
#> [6,] 13.453723
#> [7,] 4.616968
#> [8,] 3055.546022
#> [9,] 1230.535671
#> [10,] 65.559494
#> [11,] 3.833550
#> [12,] 4658.810828
det(FIM)
#> [1] 1.421054e+39
get_rse(FIM,poped.db)
#> CL V E0 EMAX EC50 d_CL
#> 3.306524 3.610227 4.575451 6.825531 30.397923 47.350220
#> d_V d_E0 d_EMAX d_EC50 SIGMA[1,1] SIGMA[2,2]
#> 56.556024 113.036005 291.542904 5854.936456 10.829134 11.705173